Molecular characterization of multidrug-resistant Escherichia coli isolated from human urine infections with their antibiogram profile
Abstract
Urinary tract infection (UTI) is the leading cause of hospitalization due to bacterial infection, and the frequency of multidrug-resistant Escherichia coli isolates from these infections is increasing worldwide. The current study aims to isolate and characterize antibiotic-resistant Escherichia coli and their antibiogram typing from urine samples of humans. From April to December 2019, a total of 60 human urine samples were collected aseptically and treated to primary isolation by propagation in nutrient broth followed by culture on various agar media. Gram’s staining, string techniques, biochemical characterization, PCR, and Sanger sequencing were performed to confirm E. coli. The Kirby-Bauer disk diffusion technique was used to test the susceptibility of all bacterial strains to thirteen typically prescribed antibiotics. The overall prevalence of E. coli in UTI was 66.67%. Three variations were noted in E. coli, all of which were single substitutions (A>T, C>T, and T>A). Phylogenetic analysis of the 16S rRNA revealed that the E. coli discovered in this study belonged to the genus Escherichia but was distinct from those identified in other countries. The antibiograms revealed that all the isolates (100%) were resistant to penicillin, ampicillin, and amoxicillin; 94.87% to doxycycline; 79.16% to gentamycin; 75.48% to ciprofloxacin; 73.07% to erythromycin; 71.66% to levofloxacin; 47.36% to ceftriaxone; and 46.66% to tetracycline. In contrast, all E. coli strains were sensitive to amikacin (95%), vancomycin (92.50%), and azithromycin (92.50%). People with a UTI often have multidrug-resistant E. coli in their urine samples, which calls for a one-health strategy to deal with this rapidly changing condition.
Keywords
INTRODUCTION
Worldwide, urinary tract infection (UTI) account for between 30 and 50% of all healthcare-associated infections and are a major public health problem [1]. Patients of all ages and both sexes may have UTI, and they can manifest in a broad variety of ways in the clinic. UTI has a global impact of over $6 billion annually, and an estimated 250 million individuals are at risk [2]. As a result of the complexity of the disease, it attacks a wide range of sensitive human body organs, including the bladder, ureters, and urethra [3]. UTI affects women 14 times more frequently than men. Due to the small size of the female urethra and its proximity to the anus, up to 40% of women might get a UTI at some point in their life, and most of these women might have recurrent UTI. In contrast, males are predisposed to infections caused by microorganisms like E. coli because their prostate glands lack the bactericidal substance and zinc necessary to effectively eliminate these threats [4]. Despite the widespread availability of medications, UTI remains one of the most common clinical outcomes. Antibiotic resistance has now become an acute ailment because of the patient’s excessive use and misuse of antibiotics without antibiogram profiling. Therefore, antibiotic resistance poses a severe public health issue in the management of patients with UTI, particularly in a developing country like Bangladesh. Finding out which bacteria cause UTI and how they respond to proposed antibiotics in clinical settings is very helpful for making empirical lines of therapy.
In fact, a UTI is caused by the prolonged growth of bacteria in the urinary system. The most common bacteria responsible for UTI in humans are Escherichia coli, Serratia marcescence, Enterococcus faecalis, Staphylococcus saprophyticus, Klebsiella pneumoniae, Pseudomonas aeruginosa, Proteus mirabilis, and Staphylococcus aureus [5]. E. coli is one of the highly diverse groups of commensal bacteria in the lower intestine of humans and animals under the family of Enterobacteriaceae, which causes a variety of infections, including UTI. E. coli bacteria are the leading cause of UTI, with 50% of hospital-acquired UTI and 85% of community-acquired UTI being caused by these bacteria [6-7]. Bacteria develop resistance to new antibiotics by changing their genetic makeup and other metabolic processes [8]. By exchanging genes for antibiotic resistance through mobile genetic components like plasmids, transposons, and integrons, antibiotic-resistant bacterial strains are constantly adapting and diversifying [9-11]. E. coli demonstrates the highest level of resistance to the most widely used and most-prescribed antibiotics [12]. In addition, the development of extended-spectrum b-lactamases (ESBLs) has sped up the global spread of E. coli resistant to extended-spectrum cephalosporins [12-13]. Because of this, it is crucial to regularly monitor whether E. coli has become resistant to currently available medicines [14]. This would help researchers figure out how E. coli became resistant and find new ways to treat UTI caused by multidrug-resistant E. coli.
In recent years, sequence analysis of the 16S ribosomal RNA (rRNA) gene has been extensively used to differentiate between strains or subspecies of many bacteria, including E. coli, based on mutations within the gene [15-19]. So, the 16S rRNA gene is the new gold standard for bacterial identification for instances of E. coli because of its remarkable degree of conservation both within and across species within the same genus [20]. So, this study was done to use molecular techniques to find E. coli in the urine of UTI patients in Bangladesh, characterize it, and find out how resistant it is to antibiotics.
MATERIALS AND METHODS
Collection and processing of samples
The Institutional Animal, Medical Ethics, Biosafety, and Biosecurity Committee (IAMEBBC) of the Institute of Biological Science (IBSc), the University of Rajshahi, approved the experimental procedures and protocols used in this study (Memo no: 56/321/IAMEBBC/IBSc). Informed consent was received from all the patients who participated in this study. Samplings were performed in April-December 2019 without any bias from a private diagnostic centre located in Rajshahi, Bangladesh. Using the standard procedure, sterile, midstream urine samples from 60 patients with UTIs were collected in wide, clean glass jars. The samples were then transported within 1 h to the Department of Veterinary and Animal Sciences, Rajshahi University, maintaining sterile and cold chain conditions for microbiological analysis. The clinicopathological features of each patient were recorded, including their age, gender, urine hue and visual appeal, blood or purulent existence, urine pH, relationship status, pregnancy status, domicile, whether they acquired their UTI in a hospital versus as an outpatient, and the season in which they developed their UTI. All methods are carried out under relevant guidelines and regulations.
Isolation and characterization of E. coli
Isolation and identification of E. coli were made by culturing on Eosin Methylene Blue (EMB), MacConkey, and Blood agar plates. Briefly, the overnight-grown broth cultures were streaked on EMB MacConkey and Blood agar plates and incubated aerobically at 37 °C overnight to observe specific colony characteristics. Single metallic sheen colonies on the EMB agar plates were indicative of E. coli. Then, Gram staining, string tests, sugar fermentation tests, methyl red tests, Voges-Proskauer tests, catalase tests, the reaction in TSI agar tests, and indole tests were used to study the colonies’ morphology and their biochemical study [21]. A UTI diagnosis was made when at least 105 colony-forming units (CFU) of E. coli per milliliter of urine were found to be present. Isolates aside from E. coli were not considered for this study.
Genomic DNA extraction
Bacterial genomic DNA was extracted from pure cultures of E. coli by the boiling method using the previously published protocol [22]. In brief, a pure colony of E. coli was put into an Eppendorf tube containing 100 μL of deionized water and gently vortexed, followed by boiling and cooling for 10 min during each step. Finally, genomic DNA is collected from each tube after centrifugation for 10 min. After ensuring the purity and concentration of the DNA using a Nanodrop Spectrophotometer (BioLab, Ipswich, MA, USA), the sample was stored at -20°C.
Polymerase chain reaction (PCR) of the 16S rRNA gene and sequencing
To confirm all E. coli isolates, PCRs targeting the E. coli 16S rRNA gene with a set of primers (Sense27F, 5′-AGAGTTTGATCMTGGCTCAG-3′ and antisense1492R, 5′-CGGTTACCTTGTTACGACTT-3′) were used [23]. A total volume of 20 µL reaction mixture for PCR reaction, which includes 10 µL of Hot Start Green Master Mix (Promega, USA), 1 µL of every ten picomoles/µL primer, 1 µL of bacterial genomic DNA at 50 ng/µL and 7 µL of nuclease-free water. Following an initial denaturation at 95°C for 3 minutes, the sample was subjected to 35 cycles of denaturation at 95°C for 30 seconds, annealing at 48°C for 30 seconds, and extension at 72°C for 90 seconds, with a final extension at 72°C for 5 minutes. The results of amplification were looked at by running them through an electrophoresis system on a 1.5% agarose gel. Visualization of amplicons was done using ethidium bromide under an ultraviolet transilluminator (Biometra, Germany). As a molecular mass DNA marker, we used the 1 KB DNA ladder (Thermo Fisher Scientific, MA, USA). Successfully amplified and specific PCR bands were cut and purified following the protocols from the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel, Bethlehem, PA, USA). The purified DNA was mixed with the primer (10–40 ng of DNA + 1 μL of 3.2 pmol primers in 10 μL of H2O) and sequenced by Sanger sequencing to further confirm the detected E. coli. Under typical cycle PCR conditions, Sanger sequencing was carried out and evaluated using an ABI PRISM 3730xl Capillary sequencer (Applied Biosystems, USA). The primers were taken out of the sequences, the sequences were aligned to make contigs with a minimum overlap of 35 bp and a minimum match percentage of 95%, and Geneious 10.2.2 (Biomatters, New Zealand) was used to make the consensus sequence.
Accession numbers for nucleotide sequences
The nucleotide sequences of the 16S rRNA gene presented in this study have been deposited to the National Centre for Biotechnology Information (NCBI, Bethesda, MD, USA) as accession numbers MT629905 and MT629906, respectively.
The construction of a phylogenetic tree
The PCR product’s base sequence was identical to those of known 16S ribosomal RNA gene sequences in the same species that were selected at random from the GenBank database. The neighbor-joining approach [24] was used to estimate the evolutionary connection of the studied bacterial isolates. The Maximum Composite Likelihood was used to find the average number of base changes per location to figure out how far apart organisms are on the evolutionary scale [25]. MEGA6 software was used to perform all the steps involved in evolutionary analysis.
The test for antimicrobial susceptibility
All 40 E. coli isolates were used for the disk diffusion test as previously described [26]. Thirteen commonly used antibiotics of various classes were employed, including fluoroquinolones (levofloxacin-5 µg, ciprofloxacin-5 µg), tetracycline (tetracycline-30 µg, doxycycline-30 µg), aminoglycosides (Amikacin-30 µg), glycopeptides (Vancomycin-30 µg), cephalosporin (ceftriaxone-30 µg), macrolides (erythromycin-15 µg, azithromycin-15 µg), aminoglycoside (gentamycin-10 µg), penicillin (penicillin-30 µg, ampicillin-25 µg and amoxicillin-30 µg). On Mueller-Hinton agar plates (Hi-Media, India), ASTs were done with newly grown bacteria that were equal to 0.5 McFarland units. Clinical and Laboratory Standards Institute (CLSI, 2016) recommendations were followed to classify the outcomes as sensitive or resistant [27]. Multidrug resistance (MDR) isolates were categorized, according to Sweeney et al., 2018 [28]. In addition, the multiple antibiotic resistances (MAR) index was calculated using the following formula:, where “a” represents the number of drugs that were resistant to a specific isolate and “b” denotes the average number of tested antibiotics [29].
Statistical analysis
Microsoft Excel 2010 was used to input all data, while IBM’s SPSS version 24 was used for analysis (Armonk, NY, USA). The rate of occurrence was determined by qualitative statistics. The fraction of antibiotic resistance among E. coli isolates and the clinicopathological characteristics were analyzed using a chi-square (χ2) test. A p-value <0.05 was judged statistically significant.
RESULTS
Prevalence of E. coli in human urine infections
Human urine samples from individuals with UTIs were tested bacteriologically, and of the 60 samples, 40 (66.67%) tested positive for the occurrence of E. coli. This was shown by isolating the organisms on a selective medium and then identifying them using Gram staining, string, and biochemical assays (Figure 1). E. coli was identified using a variety of biochemical, microscopic, and cultural techniques. Of these 40 positive cases, 6 (15%) were from males and the rest, 34 (85%), were from females (p=0.00). This finding suggests that the incidence of E. coli in UTIs was greater in female patients than in male patients. The most susceptible age group of patients to UTI was 31-45 years 16 (40%) followed by 16-30 years 11 (27.50%), 46-60 years 9 (22.50%), > 60 years 3 (7.5%), and 0-15 years 1 (2.5%) (p=0.34) (Table 1). Based on the results of this investigation, it seems that UTIs are rather frequent among adults aged 16–60 years. No statistically significant connections were found except for age between the E. coli bacterial isolates and the clinical and pathological criteria of the individuals with UTI (p>0.05).
Table 1. Percentage distribution of Escherichia coli isolated from patients with UTI according to age.
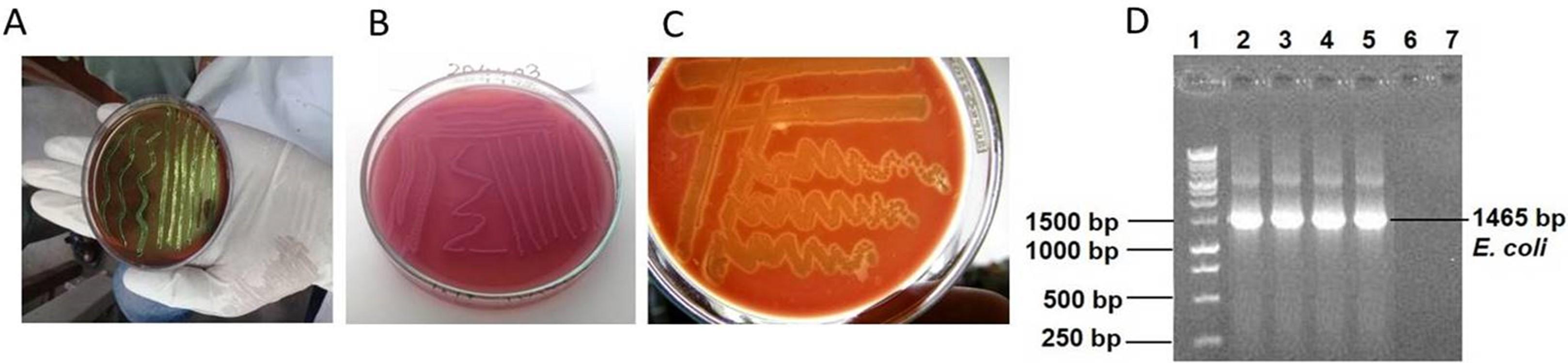
Detection of E. coli by 16S rRNA and sequencing
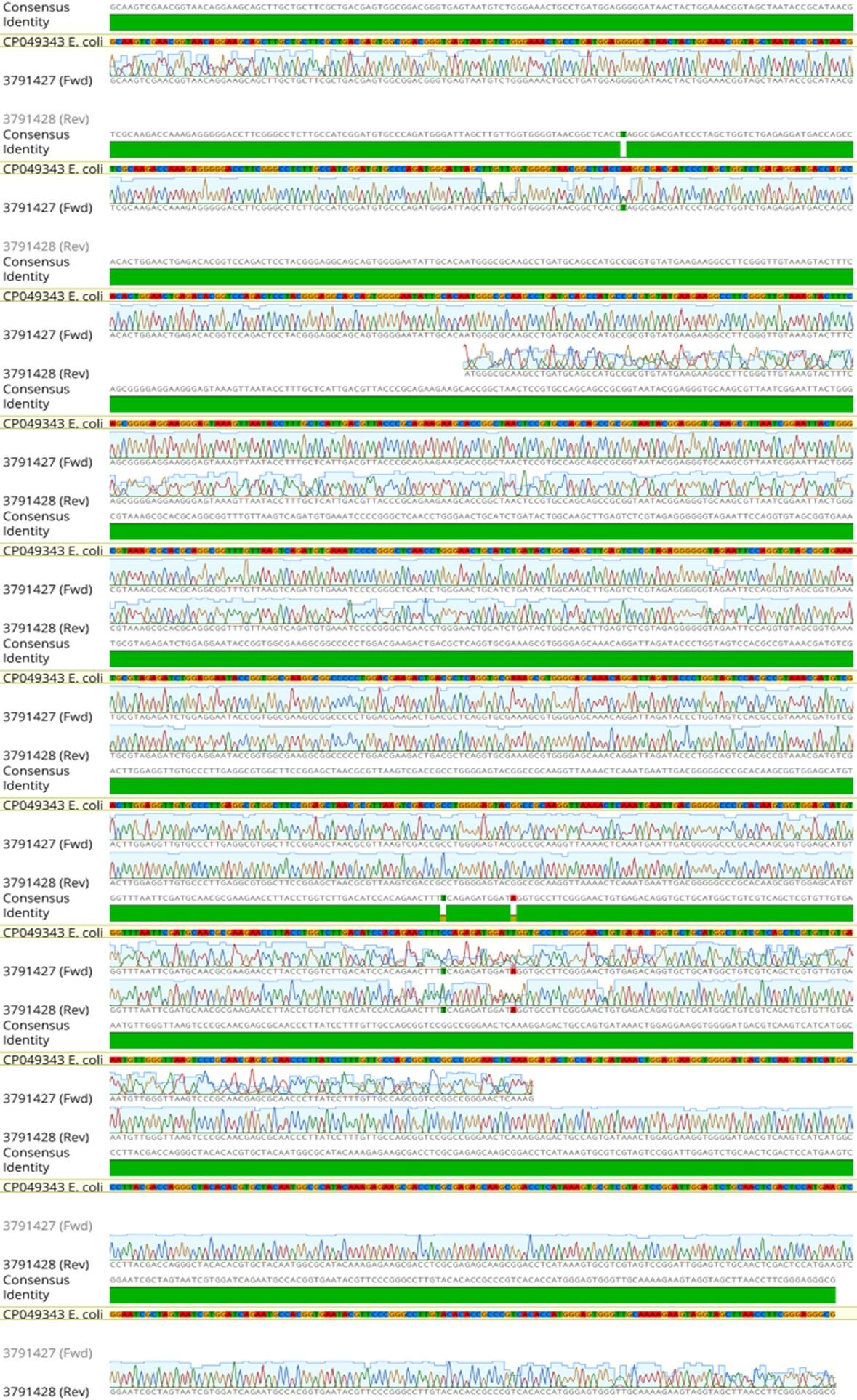
A well-established PCR approach accompanied by Sanger sequencing was used to verify all E. coli isolates detected by traditional methods. The E. coli 16S RNA gene was targeted for amplification in these samples, resulting in 1465 base pair pieces (Figure 1). Sanger sequencing showed that the chromatogram matched up with the isolation of E. coli of the PCR amplification of the 16SrRNA gene, which had 99.79% sequence similarity with an Escherichia coli strain UMB200201-11 obtained from a human sample in the USA (GenBank accession no. CP049343). There were three different new mutations discovered in E. coli, all of which were single substitutions (A>T, C>T, and T>A) (Figure 2).
Phylogenetic analysis
Phylogenetic trees were built using isolates of the same species or family from various countries to better understand the evolutionary connection of the newly sequenced 16S rRNA gene of E. coli from Bangladeshi UTI patients. Figure 3 displays the phylogenetic tree, which shows the genetic distance between the Bangladeshi isolates and those from other nations (Figure 3). Based on these comparisons, it seems that some segments of the 16S ribosomal RNA gene are translated similarly across species. A phylogenetic study shows that Bangladeshi E. coli strains are distinct from other isolates of the same species from across the world.

Antibiogram profile of E. coli isolates
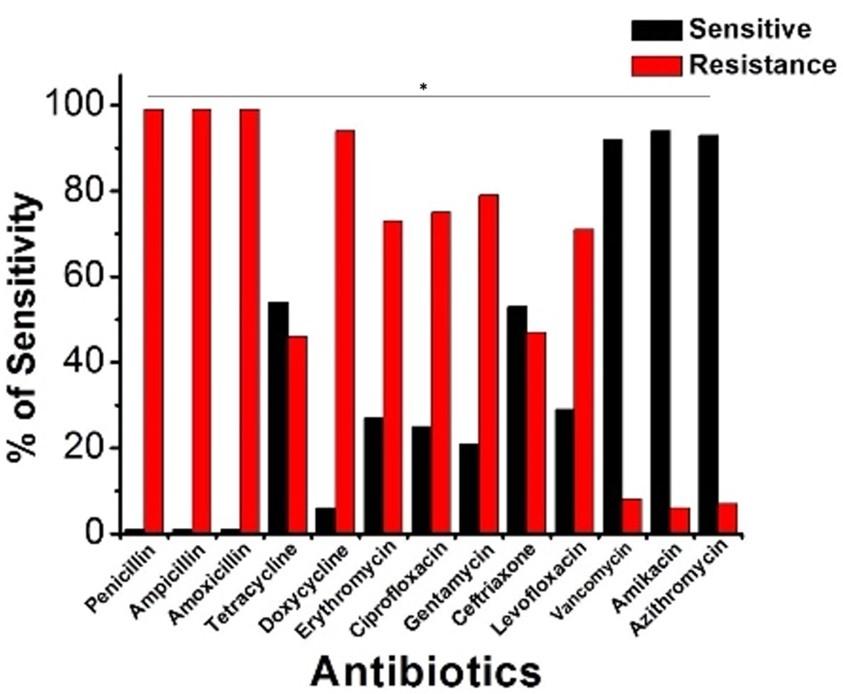
A panel of 13 medications mapped out antibiotic susceptibility using a disk diffusion test. It was shown via antibiotic susceptibility testing that all bacterial E. coli isolates examined were resistant to several antibiotics. Figure 4 displays the prevalence of E. coli resistant to several classes of antibiotics (Figure 4). All the E. coli strains examined demonstrated complete resistance to penicillin, ampicillin, and amoxicillin. Doxycycline (94.87%), gentamycin (79.16%), ciprofloxacin (75.48%), erythromycin (73.07%), levofloxacin (71.66%), ceftriaxone (47.36%), and tetracycline (46.67%) were all drugs to which a significant fraction of the isolates were resistant. Testing showed that amikacin (95%), vancomycin (92.50%), and azithromycin (92.50%) were the most effective antibiotics for treating UTI in people.
Occurrence of MDR patterns and MAR index of E. coli isolates
33 (82.50%) of the 40 E. coli isolates were MDR in phenotype. There were seven distinct resistance patterns documented in total. Among them, the highest 45.46% (15/33) E. coli isolates displayed resistance pattern no. 1 (PEN-AMP-AMX-TE-DXT-LEV-CIP-GM-E-CRO). One isolate exhibited resistance to five classes of antibiotics (seven antibiotics) (pattern no. 5). The antibiotic resistance profiling of each E. coli isolate was found to differ, with MAR indices ranging from 0.38 to 0.76 (Table 2).
Table 2. Occurrence of multidrug resistance and multiple antibiotic resistance index of E. coli isolated from Patients with UTIs.
DISCUSSION
The study’s molecular characterization and antibiogram profiling of E. coli isolates from urinary tract infections are reflective of their present prevalence. Among 60 bacteriologically analyzed UTI samples, the overall frequency of E. coli was 66.67%, which is quite close to the previous findings [28-32]. According to our study, the prevalence of E. coli in UTIs was higher (61.90%) in female patients than in male patients (38.09%). Akin to the findings of Akter et al., we observed that 64 (79%) of females and 17 (21%) of males were culturally affirming [33]. Women are at a higher risk of contracting an E. coli infection because of their short length and broader urethras; the lack of antibacterial activities of prostatic fluid; sex hormone changes that influence the mucosal adherence of bacteria; and the increased likelihood of traumatic urethral injuries sustained during sexual intercourse. UTIs, most often caused by E. coli, were more common in patients between the ages of 16 and 60. A patient’s risk of infection increased by 40% between the ages of 31 and 45 years compared to that of patients aged 46 to 60 years, and by 27.50% between the ages of 16 and 30 years compared to those aged 60 years and above. A similar percentage of patients between the ages of 37 and 47 years are susceptible to E. coli, as shown by the findings of another researcher (46.20%) [34]. The increased prevalence of UTI among people of reproductive age may be attributable to a number of factors, including sexual contact, spermicidal chemicals in contraceptives, diaphragms, menopause in women, an enlarged prostate in men, and a lack of awareness about good hygiene [35]. Findings from India and Italy [36-37] show that the prevalence of E. coli that cause UTI may depend on the age and gender of the patient.
In molecular diagnostics labs, 16S rRNA sequencing is employed in the detection of bacteria, new bacterial genera and species discovery, the detection of uncultivable bacteria, and the evaluation of culture-negative diseases [16-18, 38-39]. The comparison and alignment of the isolate’s 16S rRNA with other genus 16S rRNAs in the GenBank corroborated the results of polymerase chain reaction amplification and phylogenetic tree-building by 16S rRNA gene sequencing, which revealed that the isolate in this research belonged to the species E. coli. Additionally, we discovered three unique single-substitution mutation sites (A>T, C>T, and T>A). Species-specific bacterial detection and a non-uniform distribution of antibiotic resistance may both be attributable to mutations in the 16S ribosomal RNA gene. Because of this, the 16S ribosomal RNA gene might be used to track the source and spread of E. coli in urinary tract infections by identifying the pathogen at the species level [20].
Nowadays, UTIs are a major source of antibiotic resistance, particularly among E. coli strains. As a result, many strains exhibit resistance to many drugs [33-35]. Our study’s antibiogram of urinary pathogens shows that all E. coli isolates were 100% resistant to penicillin, ampicillin, and amoxicillin, whereas resistant patterns were doxycycline (94.87%), gentamycin (79.16%), ciprofloxacin (75.48%), erythromycin (73.07%), levofloxacin (71.66%), ceftriaxone (47.36%), and tetracycline (46.66%). This study’s findings that E. coli is highly resistant to most antibiotics are consistent with those found in Melbourne, Australia [40], Ibadan, Nigeria [41], Kanpur, India [42], Mumbai, India [43], and Kermanshah, Iran [44]. In contrast, we found that amikacin (95%), vancomycin (92.50%), and azithromycin (92.50%) were the most efficient antibiotics against E. coli for treating human UTIs. There was a high degree of sensitivity to the antibiotics meropenem (98.33%) and amikacin (90%), both of which were shown to be effective against E. coli by Sharmin, 2005 [45]. In addition, Bashir et al. found that E. coli was resistant to ampicillin, ciprofloxacin, nitrofurantoin, co-trimoxazole, and amikacin [30]. In their paper, Niranjan and Malini describe the antibiotic resistance profile of Escherichia coli in India, finding that the bacteria were resistant to ampicillin, amoxicillin-clavulanic acid, cefuroxime, ceftriaxone, and co-trimoxazole but sensitive to amikacin, piperacillin-tazobactam, nitrofurantoin, and imipenem [46].
However, as the number of treatments using these medications grows, the probability of developing resistant strains of the disease also increases. UTIs caused by MDR, and MAR bacteria pose a major threat to global public health due to the high cost of therapy and the risk of treatment failure, which can result in even death. Alarmingly, 82.50% of E. coli isolates were multidrug-resistant in nature. For instance, the MAR indices of the identified E. coli in our study ranged from 0.38 to 0.76. The MAR index suggests that antibiotics were often used to treat people in the area where E. coli was isolated, implying a significant risk source for MDR and MAR bacteria. These multi-drug resistant bacteria can potentially disseminate through the environment and transfer their resistance genes horizontally to other bacteria. When treating patients with E. coli infections, it is important that major clinics and hospitals in Bangladesh keep a continuous survey on the antibiotic susceptibility pattern and that doctors use these medicines sparingly to prevent antimicrobial resistance.
CONCLUSIONS
E. coli was found to be the most commonly isolated bacterium (66.67%) from UTIs. UTIs account for 61.9% of bacterial infections in females. Those between the ages of 16 and 30 years had the greatest UTI prevalence rate. Amikacin, vancomycin, and azithromycin were the most effective antimicrobial drugs; most of the strains were resistant to penicillin, ampicillin, amoxicillin, doxycycline, gentamycin, ciprofloxacin, erythromycin, levofloxacin, ceftriaxone, and tetracycline. This means that uropathogenic bacteria often exhibit MDR. In particular, infections of the urinary tract, particularly those caused by E. coli, have become more challenging to treat empirically as a result of widespread resistance to widely used antibiotics. Effective antibiotic prescription requires an understanding of local trends in antimicrobial resistance. People with UTIs all over the world could benefit from our research while doctors decide how to treat them.
ACKNOWLEDGEMENTS
We appreciate everyone who agreed to take part in the research. This work by SS was made possible by the Australian Research Council’s Discovery Early Career Researcher Award. The University Grants Commission of Bangladesh generously provided the study’s financing.
AUTHOR CONTRIBUTIONS
The work was designed and supervised by MHH. The research work was performed by MZI, MLM and MHH. The first draft of this manuscript was prepared by MHH and SKD. MHH and SS analyzed the data and improved the overview of the manuscript. MHH and SS critically revised, improved and approved the final version of the manuscript.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Terpstra ML, and Geerlings, SE. Urinary tract infections: how new findings create new research questions. Curr Opin Infect Dis. 2016; 29:70-72.
- [2]Mohiuddin AK. UTI prevalence among population with chronic conditions. J Med Case Rep. 2019; 1:2.
- [3]Ullah A, Shah S, Almugadam B. Prevalence of symptomatic urinary tract infections and antimicrobial susceptibility patterns of isolated uropathogens in kohat region of Pakistan. MOJ Biol Med. 2018; 3(4):85-90.
- [4]Lawhale MA, Naikwade R. Recent pattern of drug sensitivity of most commonly isolated uropathogens from Central India. Int J Res Med Sci. 2017;5(8):3631-3636.
- [5]Rahman SU, Ahmad M, Islam B, Ullah A, Rahman MU, Khan Z, et al. Isolation and identification of Escherichia coli from urine samples and their antibiotic susceptibility pattern. J Entomol Zool Stud. 2019;7(3):259-264.
- [6]Zinnah MA, Bari MR, Islam MT, Hossain MT, Rahman MT, Haque MH, Babu SAM, Ruma RP Islam MA. Characterization of Escherichia coli isolated from samples of different biological and environmental sources. Bangl J Vet Med. 2007; 5:25–32 .
- [7]Kalsoom B, Jafar K, Begum H, Munir S, ul AKBAR N, Ansari JA, et al. Patterns of antibiotic sensitivity of bacterial pathogens among urinary tract infections (UTI) patients in a Pakistani population. Afr J Microbiol Res. 2012;6(2):414-420.
- [8]Munita JM, Cesar A, Arias CA. Mechanisms of Antibiotic Resistance. Microbiol Spectr. 2016; 4(2):10.1128/ microbiolspec. VMBF-00162015. doi: 10.1128/microbiolspec.
- [9]Aminov RI. Horizontal gene exchange in environmental microbiota. Front Microbiol. 2011;2: 158.
- [10]Stokes HW, and Gillings, MR. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes in Gram‐negative pathogens. FEMS Microbiol Rev. 2011; 35:90-781.
- [11]Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin Microbiol Rev. 2018;31(4): e00088-17.
- [12]Mukherjee M, Basu S, Mukherjee SK, Majumder M. Multidrug-resistance and extended spectrum beta-lactamase production in uropathogenic E. coli which were isolated from hospitalized patients in Kolkata, India. J Clin Diagn Res. 2013; 7:449-453.
- [13]O’Neill J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. 2014, https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis.
- [14]Zinnah MA, Haque MH, Islam MT, Hossain MT, Bari MR, Babu SAM, et al. Drug sensitivity pattern of Escherichia coli isolated from samples of differentbiological and environmental sources. Bangl J Vet Med. 2008;6(1):13-18.
- [15]Magray MS, Kumar A, Rawat AK, Srivastava S. Identification of Escherichia coli through analysis of 16S rRNA and 16S-23S rRNA internal transcribed spacer region sequences. Bioinformation. 2011;6(10):370-1.
- [16]Suardana IW. Analysis of Nucleotide Sequences of the 16S rRNA Gene of Novel Escherichia coli Strains Isolated from Feces of Human and Bali Cattle. J Nucleic Acids. 2014; 2014:475754.
- [17]Johnson JS, Spakowicz DJ, Hong BY, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun, 2019;10:5029.
- [18]Ahsan N, Rahman M, Islam MN, Akhand AA. Isolation and Characterization of Multidrug Resistant Enterobacteriaceae in Urine Sample of Patients Suffering from Urinary Tract Infection with Diabetes and Nephropathy. Dhaka Univ J Pharm Sci. 2021;20(1):87-93.
- [19]Jaaz WS. Molecular detection of 16srrna gene in Escherichia coli isolated from urinary tract infection patients. Eurasia J Biosci. 2020; 14:99-104.
- [20]Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin microbiol infect. : the official publication of the European Society of Clinical Microbiology and Infectious Diseases, 2008;14(10), 908-934.
- [21]Bergey DH, Buchanan RE, Gibbons NE. Bergey’s Manual of Determinative Bacteriology, 8th ed.; American Society for Microbiology: Washington, DC, USA, 1974;966-1097.
- [22]Haque MH, Miah ML, Sarker S, Shamsuzzaman M and Shiddiky MJA. Molecular Characterization and Antibiogram Profiling of Multidrug Resistant Staphylococcus haemolyticus Isolated from Patients with Urinary Tract Infection in Bangladesh. J Bacteriol Mycol. 2021;8(2):1166.
- [23]McCabe KM, Zhang YH, Huang BL, Wagar EA, McCabe ER. Bacterial species identification after DNA amplification with a universal primer pair. Mol Genet Metab. 1999; 66:205‐211.
- [24]Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987; 4:406-425.
- [25]Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci. USA. 2004; 101:11030-11035.
- [26]Bayer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disc method. Am J Clin Pathol. 1966; 45:493-496.
- [27]CLSI. Performance Standards for Antimicrobial Susceptibility Testing: 17th Informational Supplement; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016.
- [28]Sweeney MT, Lubbers BV, Schwarz S, Watts JL. Applying definitions for multidrug resistance, extensive drug resistance and pan drug resistance to clinically significant livestock and companion animal bacterial pathogens. J Antimicrob Chemother. 2018; 73:1460-1463.
- [29]Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol. 1983;46(1):165-170.
- [30]Bashir M, Qazi J, Ahmad N, Riaz S. Diversity of urinary tract pathogens and drug resistant isolates of Escherichia coli in different age and gender groups of Pakistanis. Trop J pharm Res. 2008;7(3):1025-1031.
- [31]Jhora ST, Paul, S. Urinary Tract Infections Caused by Staphylococcus saprophyticus and their antimicrobial sensitivity pattern in Young Adult Women. Bangladesh J Med Microbiol. 2011;5(1):21-25.
- [32]Mollick S, Dasgupta T, Hasnain MJ, Ahmed M. Isolation and Characterization of Pathogens Responsible for Urinary Tract Infection in Bangladesh and Determination of their Antibiotic Susceptibility Pattern. J App Pharm Sci. 2016;6 (04):072-076.
- [33]Akter T, Mj H, Khan SA, Sultana H, Fatema K, Sa S, et al. Isolation, identification and antimicrobial susceptibility pattern analysis of Escherichia coli isolated from clinical samples of Bangladesh. Asian J biomed pharm. 2016; 6:13-16.
- [34]Alo MN, Saidu AY, Alhassan M, Mx. Prevalence and Antbiogram of Bacterial Isolates Causing Urinary Tract Infections at Federal Teaching Hospital Abakaliki I (FETHA I). Microbiol Res J Int. 2015;8(2):403-417.
- [35]Haider JS, Hasan A, Bin-Tahir K. Frequency of Urinary Tract Bacterial Infection and their Susceptibility Patterns among Hemodialysis Patients in Zliten Hospital. J Microbiol Exp. 2016; 3:00093.
- [36]Magliano E, Grazioli V, Deflorio L, Leuci AI, Mattina R, Romano P, et al. Gender and age-dependent etiology of community-acquired urinary tract infections. Sci World J. 2012;2012: 349597.
- [37]Mahajan R, Gupta S, Mahajan B. Antibiotic Susceptibility Pattern of Isolates in Urinary Tract Infection in a Tertiary Care Hospital. J Rational Pharmacother Res. 2014; 2:44-49.
- [38]Barghouthi SA. A Universal Method for the Identification of Bacteria Based on General PCR Primers. Ind J Microbiol. 2011;51(4):430-444.
- [39]Iliyasu MY, Bamanga RA, Umar AF, Agbo EB, Uba A. 16S rDNA Sequencing analysis in identification of some multidrug resistant (MDR) bacterial isolates from clinical specimens. Nig J Biotech. 2020;36(2):158-166.
- [40]Bettelheim KA, Hornitzky MA, Djordjevic SP, Kuzevski A. Antibiotic resistance among verocytotoxigenic Escherichia coli (VTEC) and non-VTEC isolated from domestic animals and humans. J Med Microbiol. 2003; 52:155-162.
- [41]Okesola AO, Aroundegbe TI. Antibiotic resistance pattern of uropathogenic Escherichia coli in South West Nigeria. Afr J Med Med Sci. 2011;40(3):235-238.
- [42]Melaku S, Kibret M, Abera B, GebreSellassie S. Antibiogram of nosocomial urinary tract infections in Felege Hiwot referral hospital, Ethiopia. Afr Health Sci. 2012;12(2):134-139.
- [43]Nerurkar A, Solanky, P, Naik, SS. Bacterial pathogens in urinary tract infection and antibiotic susceptibility pattern. J Pharm Biomed Sci. 2012;21(12):1-3.
- [44]Jalilian S, Farahani A, Mohajeri P. Antibiotic resistance in uropathogenic Escherichia coli isolated from urinary tract infections out-patients in Kermanshah. Int J Med and Public Health. 2014;4(1):75-55.
- [45]Sharmin S. Use of chromogenic media for detection of uropathogen, M phil (Microbiology) Thesis. 2005; Bangabandhu Sheikh Mujib Medical University, Bangladesh.
- [46]Niranjan V, Malini A. Antimicrobial resistance pattern in Escherichia coli causing urinary tract infection among inpatients. Indian J Med Res. 2014;139(6):945.